Microbial Strategies for Survival and Evolution
 Course Directors:
Course Directors:
Course Instructors:
Guest Lecturers (partial list):
COURSE SUBJECT
Microbial life is considerably more complex than is commonly appreciated. Far from merely proliferating exponentially until nutrients are exhausted, microbes engage in a wide range of adaptive behaviors. For example, the diauxie phenomenon associated with the nutrient induced shift of bacterial metabolism was at the heart of Monod and Jacob’s discovery of regulated gene expression. Much like cells in the multicellular organisms, microbes – bacteria and yeast for the purposes of this course – can exist in different phenotypic states. Microbes can switch between different phenotypes and the phenotypic states can persist through cell division thanks to the existence of epigenetic memory. The mechanisms of the epigenetic memory are remarkably diverse, from hysteretic transcriptional regulation circuits to metabolic circuits to DNA methylation and are a subject of much current research, focusing in particular on the role of inductive signals and stochastic fluctuations in switching between states. Interestingly, epigenetic switching coexists with mechanisms of genetic switching via genomic rearrangement (phase variation) or recurrent mutations bridging in a very interesting way the phenotypic dynamics of microbial cells and evolutionary dynamics of microbial colonies. Quantitative exploration of these phenomena will be the central themes of the course.
EXPERIMENTAL PROJECTS (preliminary)
Bet-hedging and rapid phenotypic switching
Instructors: Paul Rainey and Silvia De Monte
 in diverse and changing environments. Under predictable conditions, coordinated regulation of gene expression allows organisms to modify aspects of behavior, morphology or phenotype to match prevailing circumstances. In fluctuating, or unpredictable, environments, such strategies fail: survival requires that organisms hedge their evolutionary bets. In bacteria bet hedging is achieved by mechanisms that promote rapid stochastic phenotype switching. We will take advantage of two real-time evolution experiments using bacteria, out of which genotypes have evovled that stochastically switch between distinct cellular states. Using a combination of theory, modeling, single cell analyses and deep-sequencing we will explore cellular and mutaitonal details of these switches in order to understand evolutionary parameters affecting the success of populations. For additional background please see Beaumont et al, Nature 2009 and Libby&Rainey, Proc. R. Soc. B (2011) 278, 3574–3583
in diverse and changing environments. Under predictable conditions, coordinated regulation of gene expression allows organisms to modify aspects of behavior, morphology or phenotype to match prevailing circumstances. In fluctuating, or unpredictable, environments, such strategies fail: survival requires that organisms hedge their evolutionary bets. In bacteria bet hedging is achieved by mechanisms that promote rapid stochastic phenotype switching. We will take advantage of two real-time evolution experiments using bacteria, out of which genotypes have evovled that stochastically switch between distinct cellular states. Using a combination of theory, modeling, single cell analyses and deep-sequencing we will explore cellular and mutaitonal details of these switches in order to understand evolutionary parameters affecting the success of populations. For additional background please see Beaumont et al, Nature 2009 and Libby&Rainey, Proc. R. Soc. B (2011) 278, 3574–3583Mutational and non mutational resistance to bacteriophage.
Instructor: Josep Casadesús
Classical studies showed that bacterial resistance to phages can be acquired by mutation. In certain cases, however, bacteriophage resistance can be also acquired by non-mutational mechanisms. For instance, the Salmonella enterica opvAB operon encodes cytoplasmic membrane proteins that alter lipopolysaccharide O antigen chain length, and confer resistance to bacteriophage P22. Because expression of opvAB undergoes phase variation, S. enterica populations contain a mixture of opvAB-ON and opvAB-OFF cells. Infection of Salmonella enterica with the virulent phage P22 H5 kills the major opvAB-OFF subpopulation and selects the opvAB-ON subpopulation. Cessation of phage challenge permits the appearance of a novel opvAB-OFF subpopulation generated by opvAB phase variation. Epigenetic resistance thus pre-adapts S. enterica to survive phage challenge in a reversible manner and without the fitness costs associated with mutation. However, the S. enterica opvAB-ON subpopulation is avirulent in mice and unable to proliferate in macrophages. Phase variation of opvAB expression may thus be viewed as a tradeoff between virulence and phage resistance. We will characterize phage and bacterial populations using fluctuation tests, assays of fitness costs associated with individual mutants and flow cytometry to determine rates of switching between ON and OFF states.
____________________________________________________________________________________
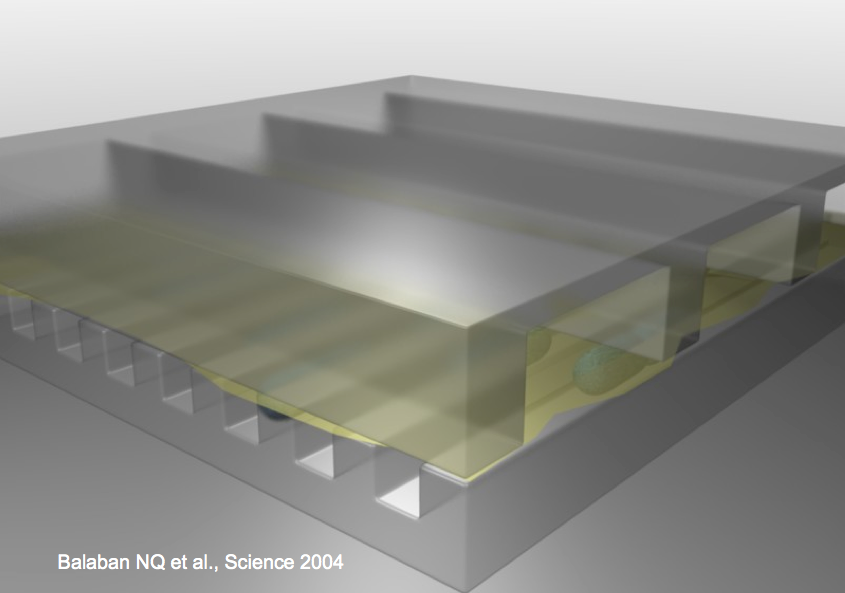
Quantitative study of “persistor” behavior
Instructor: Nathalie Questembert-Balaban
Stochasticity in phenotypic switching: Stochastic Pulse Regulation in Bacterial Stress Response
Instructor: Michael Elowitz
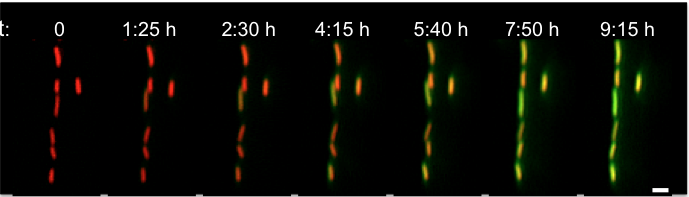
Gene regulatory circuits can use dynamic, and even stochastic, strategies to respond to environmental conditions. Using individual Bacillus subtilis cells we will explore the general stress response mediated by alternative sigma factors and characterize effects due to energy stress. Models based on minimal genetic circuitries will be constructed to which data from single cell dynamics will be fitted. The overall goal is to understand how how bacteria encode signals using stochastic pulse frequency modulation through a compact regulatory architecture.
SigmaB activation, from Locke et al, Stochastic Pulse Regulation in Bacterial Stress Response, Science 2011: Vol. 334 no. 6054 pp. 366-369
To watch a movie see http://www.elowitz.caltech.edu/graphics/1208144s1.mov
____________________________________________________________________________________
Evolution of cooperation in S. cerevisiae
Instructor: Andrew Murray
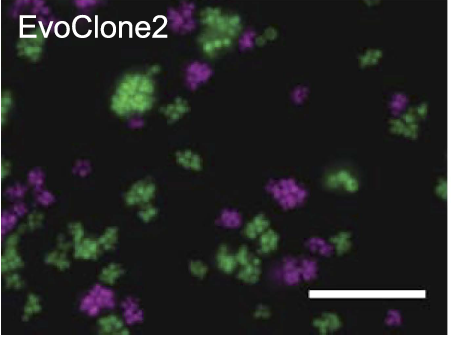
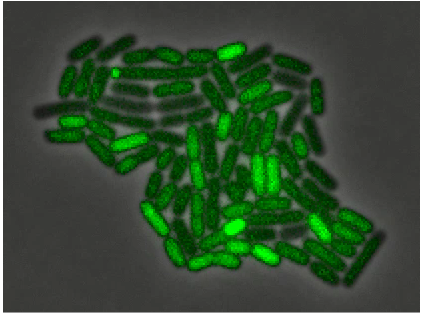
Recent experimental evolution using yeast resulted two phenotypes that depend on cells sticking together. One is the formation of multicellular clumps to successfully exploit the nutrients harvested by a secreted common good and the other is a circadian oscillator that is detected by its ability to regulate intercellular adhesion. Students will study these
strains to try and understand the mechanisms that regulate clump size. In particular we will ask how the nutrient-harvesting clumps control their size: whether connections between cells break at random or whether the probability of breaking increases with age or with clump size. Models will be developed to predict the distribution of clump sizes. For one of the nutrient-harvesting strains clump size varies depending on the nutrient it receives: wewill determine how this control works. We will also analyze the oscillator. This type requires precise temporal control of cellular adhesion. We will determine whether it is possible to monitor adhesion directly and perform experiments to unravel causal factors.
Clumpiness in the evolved clone of S. cerevisiae due to failure to separate (see Koschwanez et al eLife 2, pp. e00367 (2013)
____________________________________________________________________________________
Rapid evolution in bacterial contact inhibition system
Instructors: Sanna Koskiniemi, Christopher Hayes and David Low
Contact-dependent growth inhibition (CDI) is a phenomenon in which one bacterium inhibits the 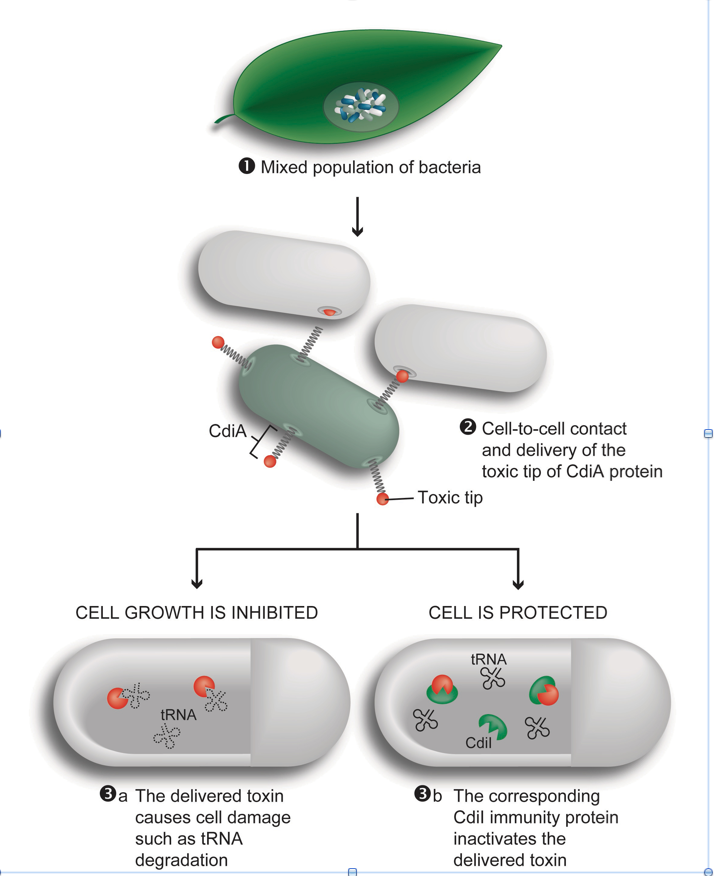 growth of another upon direct cell-to-cell contact. During CDI, a large stick like exoprotein known as CdiA with a highly divergent C-terminal end containing the toxin domain is transported to the inhibitor cell surface. Upon cell-to-cell contact the toxic C-terminus is delivered to target cells. To protect inhibition from self, a small CdiI protein binds specifically to its cognate toxin and prevents inhibition. The interactions between inhibitor and target cells are intricate and delivery of toxin to target cells requires multiple steps, including binding of a receptor on target cell surface, transfer through the inner and outer-membrane as well as binding of the toxin to an intra-cellular protein required for toxic activity. Disruption in any of these steps allows the target cells to survive and thus resistance development should occur frequently. However, the protein players identified so far consist mostly of essential proteins where even small substitutions might have large detrimental effects on the cell fitness. We have recently carried out a long-term evolution experiment using Salmonella in which derived bacteria evolved the capacity to contact-inhibit the ancestral type. We will explore the adaptive significance of this behavior through construction of simple models and will subject strains from the experiment to genetic (genomic) and phenotypic analyses in order to understand the basis of derived contact-dependent killing of ancestral types.
growth of another upon direct cell-to-cell contact. During CDI, a large stick like exoprotein known as CdiA with a highly divergent C-terminal end containing the toxin domain is transported to the inhibitor cell surface. Upon cell-to-cell contact the toxic C-terminus is delivered to target cells. To protect inhibition from self, a small CdiI protein binds specifically to its cognate toxin and prevents inhibition. The interactions between inhibitor and target cells are intricate and delivery of toxin to target cells requires multiple steps, including binding of a receptor on target cell surface, transfer through the inner and outer-membrane as well as binding of the toxin to an intra-cellular protein required for toxic activity. Disruption in any of these steps allows the target cells to survive and thus resistance development should occur frequently. However, the protein players identified so far consist mostly of essential proteins where even small substitutions might have large detrimental effects on the cell fitness. We have recently carried out a long-term evolution experiment using Salmonella in which derived bacteria evolved the capacity to contact-inhibit the ancestral type. We will explore the adaptive significance of this behavior through construction of simple models and will subject strains from the experiment to genetic (genomic) and phenotypic analyses in order to understand the basis of derived contact-dependent killing of ancestral types.
For additional background please see Hayes et al, Cold Spring Harb Perspect Med 2014
