
Application deadline is March 1, 2015.
Course Directors
Course Instructors
Mónica Bettencourt-Dias (Inst. Gulbenkian de Ciência), Cassandra Extavour (Harvard University), Michael Lynch (Indiana University), Jitu Mayor (NCBS Bangalore), Daniel Needleman (Harvard University), and José Pereira-Leal (Inst. Gulbenkian de Ciência)
Guest Lecturers (partial list)
Course subject
Recent years have witnessed a revolution in our ability to quantitatively query and manipulate biological systems. Live imaging and genome editing techniques combined with quantitative approaches have made it possible to uncover the molecular basis of many cellular processes. Indeed, cell biology has made tremendous progress towards revealing the molecular mechanisms involved in a plethora of processes ranging from cellular maintenance to the coordinated cellular decisions involved in embryonic development. However, these studies on how cellular functions are realized have been mostly devoid of the question of how these same functions came to be through evolution and natural selection.
The quantitative study of evolution, on the other hand, has been primarily focused on a description of evolutionary dynamics at the scale of populations. Here, the evolutionary history of species in a population is quantified based on the natural selection afforded by pressures such as competition with other species and changing environmental conditions. These studies focus mostly on the effect of mutations on fitness but not on how these mutations, through their effect on cellular function, lead to a reshaping of evolutionary landscapes.
A gap has existed between cell and evolutionary biology. Namely, the uncovering of how novel cell biological function evolves as organisms adapt to different environmental conditions. The time is ripe to bridge this gap as the technology that has allowed for the cell biology revolution in model organisms can now be applied in the context of their fully sequenced non-model organism counterparts. The aim of this course is to make progress towards filling this gap by focusing on the evolution of cellular processes ranging from horizontal gene transfer, to the evolution of intracellular functions such as trafficking and mitosis, to the evolution of embryonic developmental programs. The course will combine state-of-the-art in vivo microscopy, bioinformatics analysis and theoretical modelling in a plethora of model and non-model systems aimed at uncovering the evolutionary basis of cellular function and behavior at these multiple scales.
Course structure
This five week course will consist of a first "bootcamp" week to be followed by two 2-week research project sessions. Each session will consist of three or four experimental threads led by instructors and TAs who will closely work with groups of 4-5 students. Each day will consist of a morning lecture and discussions followed by lab work late into the evening.
The bootcamp week will be aimed at introducing all students to a basic set of microscopy, quantitative image analysis using Matlab and theoretical modeling that will be necessary for the later research projects. The lectures will take place in parallel with the KITP workshop "Evolutionary Cell Biology" providing a fertile forum for discussions and the exchange of ideas with the workshop participants.
EXPERIMENTAL PROJECTS (preliminary)
The Evolutionary Cell Biology of Gene Transfer. Instructor: Rob Phillips.
Microbial populations live in a fertile soup of foreign genetic information. Whether by viral infections, conjugative transfer, or direct uptake of the DNA through transformation, bacteria frequently share genetic information with their neighbors through horizontal gene transfer. Despite its deep evolutionary importance, the mechanistic details about how such gene transfer works (both at the molecular and systematic level) remain enigmatic. In our laboratory rotation, we will use microscopy to systematically explore how bacteria take up DNA. By using fluorescently labeled proteins that bind to specific sequences on the transferred DNA, we will develop a real-time picture of DNA uptake and dynamics and consider its effect on evolution across many timescales.
Vesicle Transport Evolution in the Ciliated Protozoan Paramecium. Instructor: Michael Lynch.
In eukaryotic membrane trafficking, both soluble cargo and membrane-bound components must travel efficiently and specifically between organelles via lipid-bound vesicles. These vesicles are directed specifically to their target membranes via sets of interacting protein determinants that are themselves tightly controlled in space and time. These protein determinants are members of large gene families, whose duplication and diversification have enabled trafficking pathways in different eukaryotic lineages to diversify and take on new roles. We will focus on one trafficking pathway, endocytosis, whose associated protein determinants have shown remarkable plasticity over evolutionary time. We will identify the molecular determinants involved in one endocytic trafficking step, trace their evolutionary relationships, and then track them at the subcellular level. We will look particularly for instances of functional change, or hints of functional diversification occurring in these determinant subfamilies, and test our hypotheses of functional change gleaned from targeted sequence analysis.
This work will employ members of the Paramecium “aurelia” complex, a cryptic species complex of ciliates that diverged several hundreds of millions of years ago following two complete genome duplications. As outgroup comparators, several pre-duplication species are available. Having a large macronucleus, ciliates are uniquely amenable to rapid transformation by injection of chromosomal constructs with key genes fused to GFP, which can then be used to visualize subcellular localizations of gene products.
The evolution of gene regulation and pattern formation in development. Instructor: Hernan Garcia.
The evolution of embryonic development is the substrate for the emergence of new body plans. The general features of these developmental programs are highly conserved between closely-related species. Indeed, in the Drosophila family, of which the fruit fly D. melanogaster is its most prominent member, the same transcription factors lead to the expression of genes in similar patterns in the early embryo. Despite phenotypic similarities in developmental programs, homologous regulatory decisions are realized by different arrangements of binding sites for repressors and activators in each species. For example, the regulatory region leading to the famed stripe two of the eve gene has endured significant rearrangements as binding sites have been deleted, appeared or changed in their affinity and in their spacing. Still, all of these regulatory regions lead to stripes that are qualitatively comparable and that can even rescue development when inserted into different species.
Using novel imaging techniques that allow for the quantification of transcription in single cells in living embryos we will determine how different species create the “same” pattern of gene expression. We will insert regulatory regions for the eve stripe 2 from different species as well as proposed ancestral reconstructions into D. melanogaster and some of its allied species. These experiments will open the door to determining the role of regulatory drift in these highly conserved programs as well as generate a “phylogeny of strategies for pattern formation”. This phylogeny will then be compared to the more familiar phylogeny of the DNA regulatory sequence in order to develop theoretical models of how regulatory architecture determines function and how both evolve in tandem. Finally, in order to explore the limits of what evolution could have created we will use synthetic biology to rewire this network by creating the same stripe in response to engineered, exogenous transcription factors and comparing the dynamics of formation of this genetic rescue to those dynamics of the wild-type stripes.
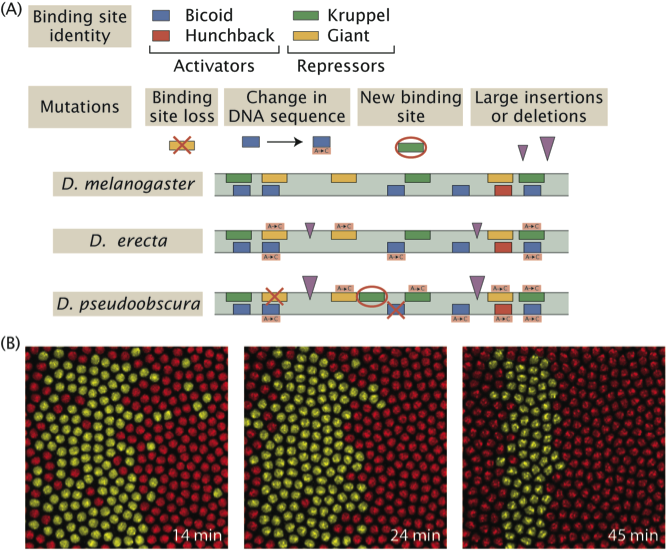
Evolution of pattern formation strategies. (A) Different fly species realize a stripe of expression of the same gene by using different regulatory architectures. Namely, the placement, number and affinity for activators and repressors in the vicinity of the gene. (B) Live imaging techniques give access to the dynamics of stripe formation in different species and to contrasting this “phylogeny of pattern formation strategies” to the more familiar DNA sequence phylogeny.
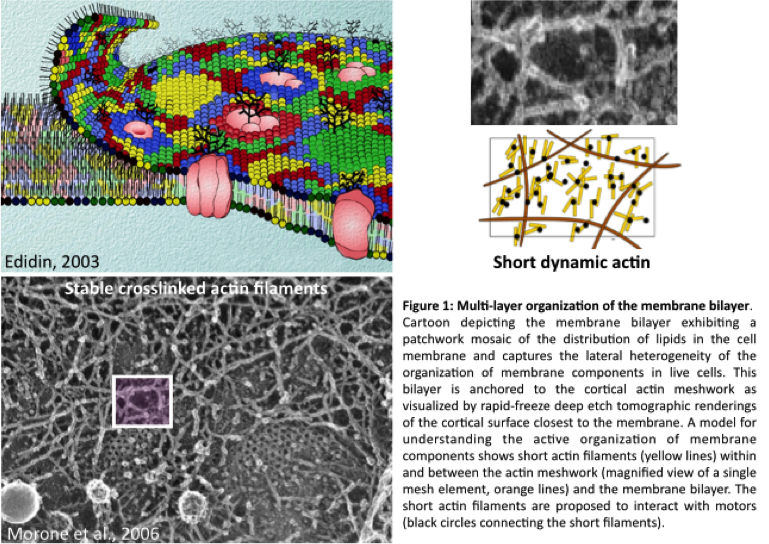
The evolution of an active membrane composite. Instructor: Jitu Mayor with Thomas van Zanten
The outer membrane of the living cell is the interface that demarcates the cell and its environment. Its chemistry is largely build up from lipids and proteins. Local organization of this chemistry primes the cell to adapt and react to the external milieu and communicate information about its internal state. In a range of metazoan cells the membrane organization is not simply passively constituted, but it is actively maintained away from thermodynamic equilibrium. This characteristic emerges from an interplay between the constituents of the membrane closely juxtaposed to a dynamic actin-based cortical layer, together forming an active composite. Cells consume energy to mechanically restructure this composite that consists out of, amongst others, the evolutionary conserved proteins such as actin, actin binding proteins and myosins. Perturbing the non-equilibrium state of cells has a dramatic effect on essential functions such adhesion or endocytosis.
Do the different cellular organisms we find in the local marine or soil environment display an active organised plasma membrane? Using an experimental scheme that was originally used for studying the membranes of metazoan cells we will explore the organisational capacity of membranes across the phylogenetic tree. This approach will consist of doping the membranes of protists with fluorescence lipid dyes and imaging FRET between the doped lipid probes to evaluate their local organization at the nanometer scale. Another way to explore this question is take a more bio-informatics approach and trace back different proteins over evolutionary time using available genome data.

Combined projects of Monica Bettencourt-Dias, Cassandra Extavour, Jose Pereira Leal, and Dan Needleman.
Within-Species Variation in Cell Biology in Drosophila
Everyone is unique. We have different appearances, different behaviors, and different talents. But to what extent are the dynamics and organization of our cells different? The answer is currently unknown. Variations in cellular behaviors between different people likely underlie many of our obvious external differences and might be responsible for differences in susceptibility to disease. Understanding the extent and nature of the genetic basis of within species variations in cell biology will also help to understand the evolution of variations in cell biology.
Recent work in the nematode Caenorhabditis elegans has shown that there is extensive within-species variation for the structure and dynamics of the mitotic spindle. We will extend this work by investigating the extent of within-species variation for a variety of cell biological features in the fruit fly Drosophila melanogaster. We will use quantitative approaches to characterize this variation and we will attempt to map the genetic basis of any differences we discover.
Between-Species Variation in Cell Biology in Insects
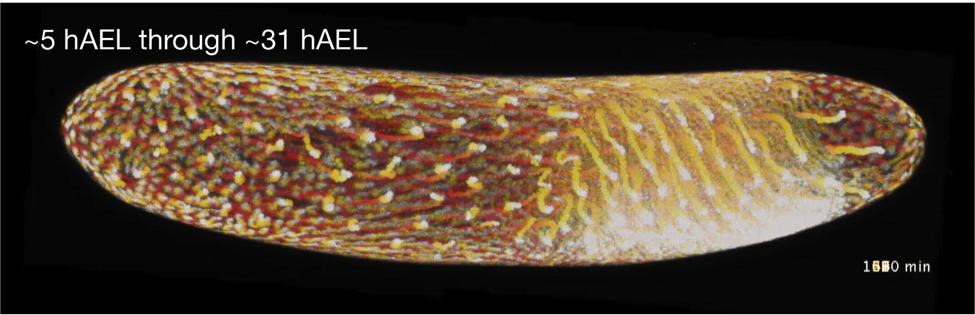
Above is a “t-projection” of a 26-hour whole Gryllus (cricket) embryo time lapse of the H2B-GFP cricket transgenic line, taken from 5 to 31 hours after egg laying (AEL). Each time point is colored in a different color along a gradient lookup table and then stacked (with dark warm colors at the beginning and yellow to white at the end of the time lapse). This shows the general course of the time lapse. (Taro Nakamura & Seth Donoughe)
Drosophila melanogaster has served as the exemplar for molecular and quantitative understanding of insect development for nearly half a century. However, this fly belongs to the lineage furthest derived from the last common insect ancestor, and its developmental and cell biological characteristics are unlikely to be representative of the most species-rich animal group, the insects. Working with not only Drosophila, but also the beetle Tribolium castaneum and the cricket Gryllus bimaculatus, students will be able to perform cell biological comparisons between lineages that diverged hundreds of millions of years ago, yet still share a highly conserved adult body plan.
Because early insect embryos undergo syncytial cleavage, in which nuclear cleavage takes place without cytokinesis, these embryos are essentially huge multinucleate cells. Moreover, although all of these insects reproduce sexually, their haploid, unfertilized eggs can be induced to begin cleavage and start to proceed through development. This provides an opportunity to compare the dynamics of nuclear spacing, synchrony and number of nuclear divisions, sexual versus asexual development, and nuclear dynamics across a broad phylogenetic range.
For both Tribolium and Gryllus, we will provide stable transgenic lines expressing fluorescently labeled cell components, enabling live imaging and quantitative analysis of image data. In addition, we will be able to manipulate embryogenesis in these animals by injecting mRNAs encoding proteins of interest, including fluorescent tags for cell biological components, pharmacological agents, and inducing RNA interference against genes of interest.

A zoomed in view of nuclei at the mid-blastoderm stage of Gryllus, with four successive time points 10 minutes apart superimposed in different colors. For some nuclei, there is no overlap between neighboring time points. (Seth Donoughe)
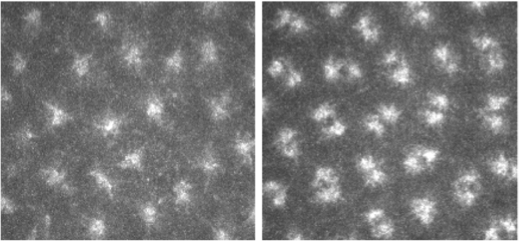
Visualization of centrosome duplication using labelled tubulin in embryos.
Cellular Evolution In Cancer- Does It Provide New Therapeutic Opportunities?
Cancer cells show several modifications in cellular characteristics, that allow them to survive, proliferate and conquer new environments. Those modifications are often specific of cancer cells and can be used for selectively killing them.
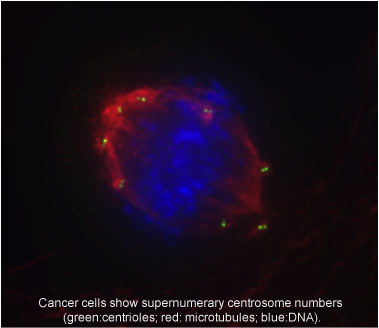 The centrosome, the major microtubule-organizing center in animal cells, participates in processes such as cell division, migration and establishment of cell polarity. Centrosome aberrations are commonly observed in cancer. Those changes might lead to increased genomic diversity and ability to invade and conquer new environments. However, those changes can also lead to very abnormal divisions that result in cell death. Mechanisms that allow cancer cells to survive in the presence of centrosome aberrations have been recently described and are being used to selectively kill cancer.
The centrosome, the major microtubule-organizing center in animal cells, participates in processes such as cell division, migration and establishment of cell polarity. Centrosome aberrations are commonly observed in cancer. Those changes might lead to increased genomic diversity and ability to invade and conquer new environments. However, those changes can also lead to very abnormal divisions that result in cell death. Mechanisms that allow cancer cells to survive in the presence of centrosome aberrations have been recently described and are being used to selectively kill cancer.
We have recently conducted a phenotypic screen for centrosome changes in a panel of 60 human cell lines that reflect different tumor types. We have seen that centrosome aberrations are a hallmark of cancer cells. While there are a few major mechanisms used to cope with those changes, we have observed that cancer cells “are creative” and can adapt in multiple ways. We will extend this work by investigating the strategies cancer cells use to adapt to centrosome changes. We will do so by using quantitative analyses of cell cycle progression and mitotic division, while monitoring chromosomes, mitotic spindle and centrosomes in live cell populations. Information on gene expression and proteome is available for the different cell lines, so ultimately cellular mechanisms might be linked to molecules. We can further investigate whether mechanisms of adaptation are already present within the population as standing variation, or arise after the appearance of centrosome changes. We will do that by acutely manipulating centrosome and spindle structural properties in normal and cancer cells, and investigating how they adapt to them.
We can also model those processes to predict whether drugs that target them would be effective in eliminating cancer cells. This project will involve microscopy, tissue culture, genetic manipulation of cells, quantitative imaging, bionformatic analysis and modeling.
Natural variation in the cell biology of marine micro-organisms
Life started in the sea, and the sea is still a source of breathtaking cellular diversity. This includes serial endosymbiotic systems where we find stable associations of cells nested within cells. For example, some dinoflagelates are found in algal blooms, where they co-exist alongside cells that have a silica-based cell-wall with amazing diversity and beauty under the microscope. The two types of organisms are examples of microalgae, primary biomass and oxygen producers in the ocean. They include many toxic species that experience explosive growths, giving rise to algal blooms that cause untold economic costs to a variety of industries worldwide, for example including shellfish farming and fisheries.
Marine micro-organisms are a good system to study natural variation of cells “in the wild,” particularly algal blooms where high concentration of diverse species co-exist. While a lot of attention has been devoted to inter-species variation and to species composition of marine micro-algae, little attention has been devoted to within-species variation. For example, do all individuals in a population for a given species multiply at the same rate? This is an important cell biology question with an impact in the dynamic of algal blooms.
In this project we will sample the local marine environment and characterize its species diversity using different techniques. We will isolate species and expand them in culture for cell biological characterization, studying natural variation of division rates, cell size, etc. We will pay particular attention to characterizing microtubule-derived organelles and structures, such as centrosomes and mitotic spindles, as proposed in the other projects in this session.
We will also explore the genetic variation and the basis of phenotypic variation using a genomics and bioinformatics approach. We will analyze publicly available genome sequence data, using bioinformatics and molecular evolution approaches, to detect for signatures of selection, expansion of protein families, variation of biophysical properties of proteins etc., in relation to phenotypic variation.
Students will be able to choose to focus more on the computational or on the wet-lab side of the project, or a hybrid of both.
