Application: deadline March 10, 2019
Letters of recommendation: two per applicant, deadline March 10, 2019
Course Directors:
Instructors:
Dominique Bergmann (Stanford), Rich Carthew (Northwestern), Mathieu Coppey (Inst. Curie), Liam Dolan (Oxford), Kenneth Irvine (Rutgers), Madhav Mani (Northwestern), Adam Martin (MIT), and Max Wilson (UCSB)
Guest Lecturers (partial list):
The summer course is closely linked to the concurrent KITP program Morphogenesis in Animals and Plants: Search for Principles. Course participants will attend the program's daily research seminars as part of the course curriculum. Students and lecturers will also have frequent opportunities for less formal interactions. Confirmed program participants include A. Boudaoud (ENS Lyon), M. Bagnat (Duke), J. Briscoe (Crick Inst.), C. Extavour (Harvard), C. -P. Heisenberg (IST Austria), M. Heisler (U. of Sydney), F. Jülicher (MPI-PKS), T. Lecuit (Collège de France and IBDM Marseille), O. Leyser (Cambridge), E. Meyerowitz (Caltech), D. Montell (UCSB), N. Nakayama (U. of Edinburgh), J. Nemhauser (U. of Washington), A. Roeder (Cornell), E. Sharon (Hebrew U.), S. Shvartsman (Princeton), A. Shyer (Rockefeller), E. Siggia (Rockefeller), X. Trepat (IBEC), and A. Warmflash (Rice).
Course Subject
Morphogenesis is the process by which a single cell transforms into a complex organism. During development, cell growth, division, and differentiation are tightly controlled to form properly shaped, functioning organs. This remarkable control over form, fate, and function is coordinated by a tight interplay of the genetic program of development and physical interactions between cells.
Coordination of physical processes is equally challenging in the development of animals and plants, but the challenge takes different forms on account of the differences in the structure and organization of their cells and tissues. Thus each kingdom developed independent solutions to the same problem of morphogenesis: encoding of shape and structure of a multicellular organism in the genetic code and the process of development. This course will provide a broad and synthetic view of the subject, aiming to distill the general ideas and principles of development from the diversity of morphogenetic processes.
This hands-on research course will integrate laboratory projects and mathematical modeling, aiming to achieve quantitative understanding of specific morphogenetic processes in a variety of model developmental systems including the fruit fly, the liverwort (Marchantia), and tissue culture. It will emphasize quantitative questions in experimental design and new techniques like optogenetics and in toto microscopy, along with advanced image analysis and mathematical model building.
Course Structure
The course is 5 weeks, beginning with a one-week bootcamp, followed by two 2-week lab sessions. Each session will consist of about four experimental projects led by instructors and TAs working closely with groups of 4-5 students. The daily routine involves two morning lectures and discussions, followed by lab work in the afternoons until late in the evenings.
During the bootcamp, students will acquire skills needed to succeed in research projects, including microscopy, quantitative image analysis, and theoretical modeling. There will also be an introduction to model systems of development and laboratory techniques.
The morning lectures will take place in the context of the parallel running KITP program “Morphogenesis in Animals and Plants: Search for Principles”, providing an excellent setting for discussion and exchange with program participants. The scientific agenda of the program aims to closely study morphogenesis in animals and plants, bringing together these diverse communities with experts in physical modeling of morphogenesis.
The program will study how common physical challenges, experienced by animals and plants are solved. Plant and animal lineages diverged before the evolution of multicellularity, strikingly manifested at the molecular level, where each kingdom separately optimized genetic pathways.
Specifically, the program will seek to develop answers towards these questions:
- How are cell behaviors coordinated during development over long ranges?
- How are growing organs divided into distinct zones?
- Are there common organizing principles underpinning regulation of these processes across kingdoms?
- Is there mechanical regulation of growth?
Experimental Projects (partial list)

Max Wilson: Virtual morphogenesis using optogenetics
Cells have evolved to survive in fluctuating environments by constantly sensing and responding to external stimuli. Signal transduction pathways are the crucial link through which cells transmit information about their environment to the genome, where they can coordinate a response. In this way, cells create an internal representation of the external world, which they use as the logical basis for executing complex behaviors.
Students will explore the how induced pluripotent stem cells (iPSCs) process spatially and temporally encoded information to make cell fate decisions. They will learn how to engineer light-based control over a number of important signaling pathways in development. They will then be provided with iPSCs in which one of their signaling pathways has been pre-engineered to be controlled with light. Students will spend the rest of the course designing experiments using state-of-the-art light delivery devices to explore a developmental fate choice of interest and developing models to characterize the decisions making apparatus in iPSCs. We hope that students come away from this course with a practical understanding of optogenetics, the various ways in which light can be delivered to cells, and a notion for how signaling information is encoded/decoded by iPSCs.
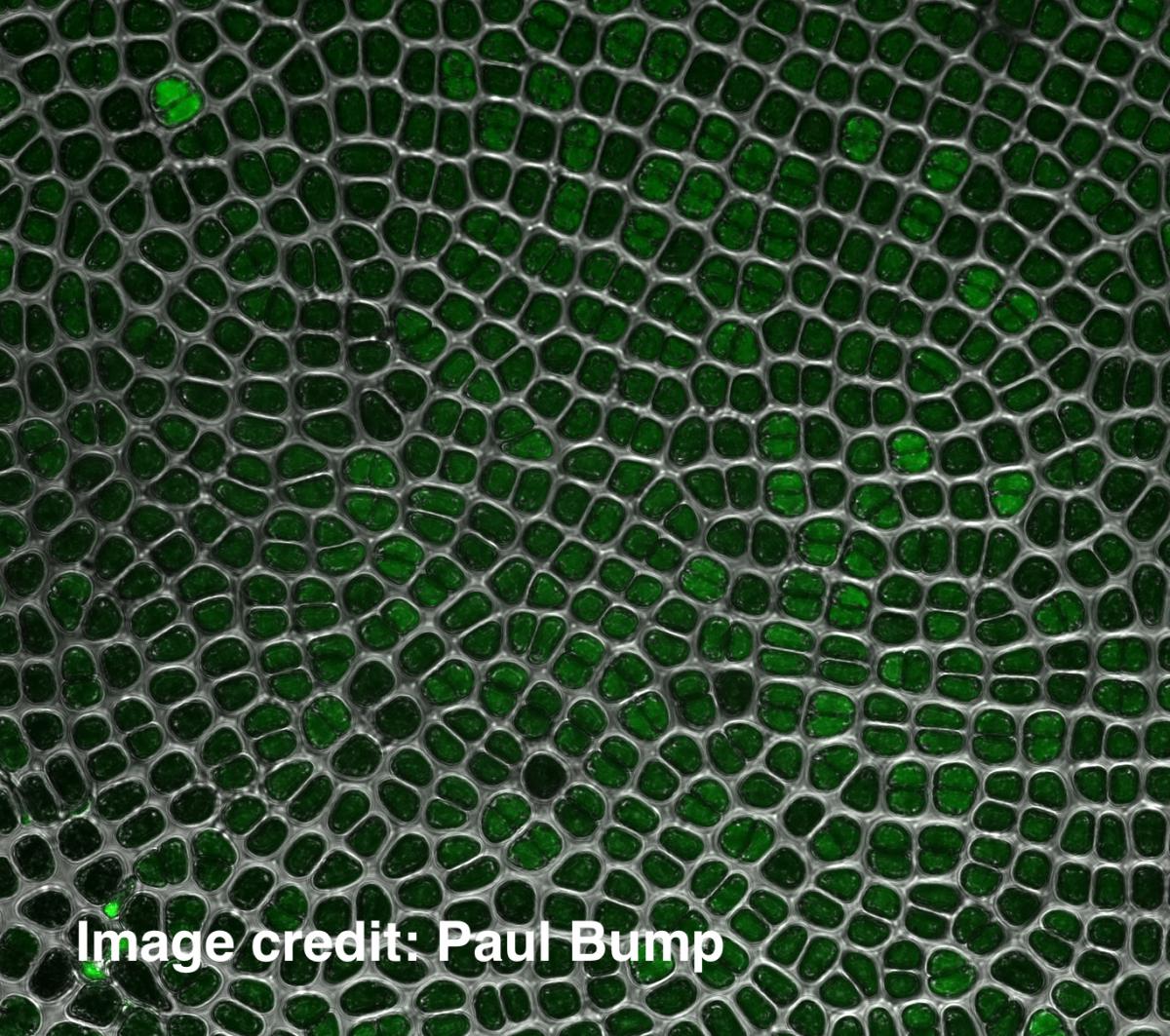
Dominique Bergmann: Pattern formation in walled cells of plants and algae
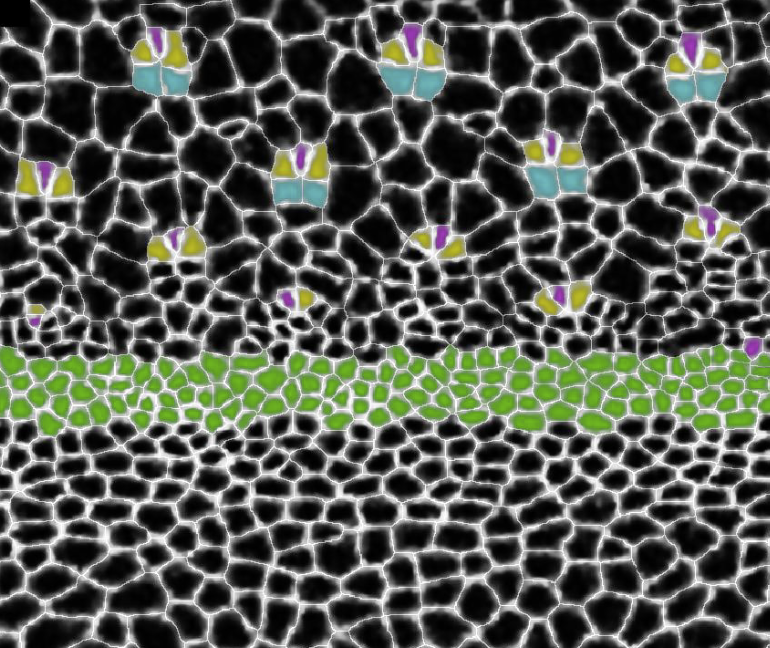
Rich Carthew and Madhav Mani: Pattern formation in the Drosophila eye imaginal disc
Mathieu Coppey: Optogenetic control of collective cell migration
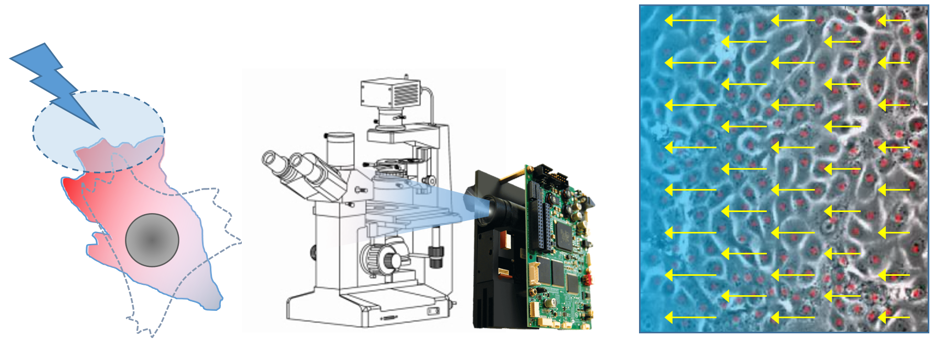
Critical events happening during morphogenesis require the collective migration of hundreds or even thousands of cells. Such collective movements depend on two main ingredients. First, physico-chemical signals of large spatial extent, such as morphogenetic gradients or mechanical stresses, inform and guide migration. Second, the communication between cells, via secreted molecules or direct interactions, ensures the coordination and self-organization of the collective migration.
In this experimental project, to evaluate the respective roles of these two ingredients, we propose a quantitative experimental approach based on the light control of long-range guidance cues. The principle will be to actuate collective migration thanks to light patterns shined on engineered cells in culture. Cells will express blue light sensitive molecular systems that can activate intracellular signaling pathways (MAPK and RhoGTPases) known to be involved in collective cell migration. The patterns of light will be generated using a micro-mirror system (DMD) coupled to an epifluorescence microscope. The collective movement will be analyzed either by tracking individual cell trajectories or using image processing methods (PIV, optical flow).
Kenneth Irvine: Three dimensional analysis of morphogenesis in imaginal discs
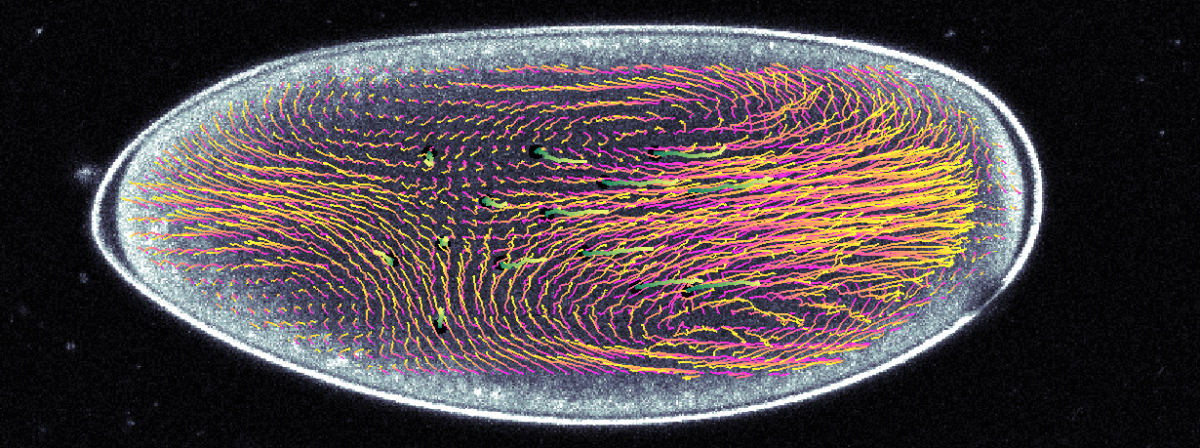
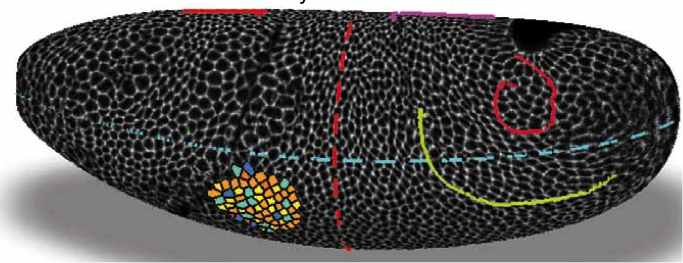
Adam Martin and Sebastian Streichan: Cellular flow in Drosophila embryo
Stefano Di Talia: Cytoplasmic flows and morphogenesis of early Drosophila embryos