This course was postponed to summer 2020 due to the global coronavirus pandemic.
Course Directors:
Instructors:
David Baltrus (U. of Arizona), Mike Boots (UC Berkeley), Jonathan Friedman (Hebrew U.), Britt Koskella (UC Berkeley), Itzik Mizrahi (Ben-Gurion U.), and Alex Petroff (Clark U.)
Guest Lecturers (preliminary list):
The summer course is closely linked to the concurrent KITP program The Ecology and Evolution of Microbial Communities. Course participants will attend the program's daily research seminars as part of the course curriculum. Students and lecturers will also have frequent opportunities for less formal interactions. Confirmed program participants include Joy Bergelson (U. Chicago), Devaki Bhaya (Carnegie Inst.), Michael Doebeli (UBC), Daniel Fisher (Stanford), Susan Holmes (Stanford), Terry Hwa (UCSD), Jay Lennon (Indiana U.), Pankaj Mehta (Boston U.), Nancy Moran (UT Austin), Andrew Murray (Harvard), Victoria Orphan (Caltech), Paul Rainey (Max Planck), Alvaro Sanchez (Yale), Daniel Segrè (Boston U.), Kim Sneppen (Bohr Inst.), Alfred Spormann (Stanford), Ned Wingreen (Princeton)
Course Subject
Microbes form diverse and complex communities in both environmental and host-associated niches. These communities are ubiquitous and have global impacts on processes as diverse as biogeochemical nutrient cycling in terrestrial and marine ecosystems; the formation of intimate associations with varied hosts, and the suppression or emergence of disease. This hands-on research course will integrate laboratory projects and mathematical modelling to provide students with an understanding of the new tools and analytical approaches used to address questions in microbiome ecology and evolution. Students will gain experience developing models and reproducible analysis workflows to ask sharp, quantitative research questions focused on the discovery of the principles underlying microbial community structure, function and evolution.
Course Structure
The course is 5 weeks long, and runs concurrently with the KITP Program on the Ecology and Evolution of Microbial Communities. The course begins with a one-week bootcamp on basic wet and dry lab techniques. Students will learn how to develop and implement workflows for reproducible and scalable data analyses and learn about model development, analysis, and numerical solving techniques. Students will then break into project groups. A selection of 2 and 4-week projects are available on a variety of topics, with up to four projects running concurrently at one time. The experimental projects are led by instructors and TAs working closely with groups of 4-5 students. Additional guest lectures and training modules will be provided throughout the course for students to learn the basics of microbial metabolism, environmental microbiology, disease ecology and eco-evolutionary theory. Students complete the course with short presentations on their project findings.
Students regularly attend the two morning lectures offered as part of the program, followed by lab work in the afternoon and evening. Students are welcome to join visiting lecturers and program participants for tea breaks and evening BBQs at the Munger Physics Residence. Weekends are open for students to explore and enjoy the many recreational opportunities in the Santa Barbara area.
No tuition or lab fees. Admitted students are provided with lodging and meals.
Experimental Projects (partial list)
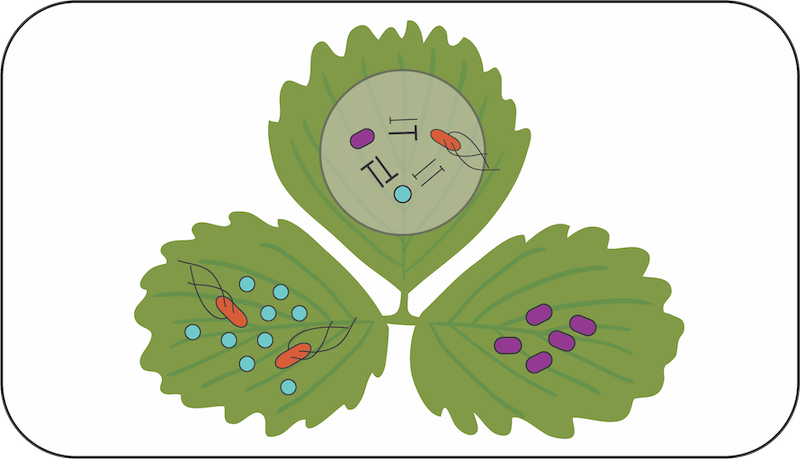
Impact of toxin-mediated competition on phyllosphere community structure
David Baltrus
Computing community function in polysaccharide-degrading bacteria
Otto Cordero, Seppe Kuehn, Itzik Mizrahi
How do we “compute” the function of a community if we know the function of its parts? We will focus on polysaccharide-degrading bacteria from the ocean, in particular those that consume chitin or alginate. These organisms are in culture and belong to genera like Vibrio and Alteromonas. We will define “function” as the respiration rate, polysaccharide consumption and biomass production rates. Respiration will be measured using pressure sensors that quantify oxygen consumption. Students will use this tool to measure respiration rates of mono- and co-cultures to determine whether community function is some linear combination of the function of the parts or whether one needs to account for higher order effects. We will build simple quantitative models to define the expected outcome of cocultures simply assuming that respiration rates are proportional to growth kinetics. Models will be “updated” with experimental observations. Because growth kinetics can be cooperative (rates can be density dependent), students will also have to study how the function of the co-cultures depends on initial conditions (fraction and concentration of each species, as well as substrate concentration).
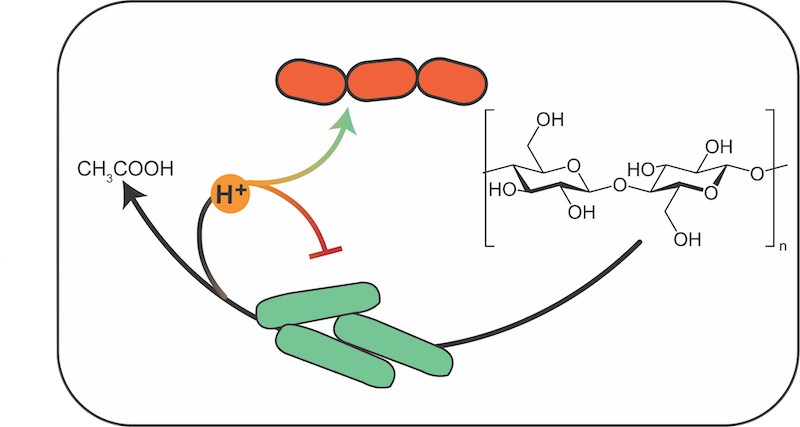
Syntrophic ecological circuits in anaerobic systems
Otto Cordero, Seppe Kuehn, Itzik Mizrahi
Cellulose hydrolysis has been shown to increase in the presence of methanogens that serve as hydrogen sinks. In this system, a fiber degrader such as Fibrobacter succinogenes breaks down cellulose but is inhibited by its own metabolic waste, namely H+. Methanogens like Methanobrevibacter smithii consume hydrogen and decrease the ∆G of the fermentation reaction. This is the quintessential form of syntrophy in anaerobic systems. These types of reactions have been studied for many years by engineers, but simple (physics inspired) models that capture different dynamic regimes and phase space are lacking. Students will start by building a naïve model of the system based on literature. These models are based on Monod growth kinetics with rates that are adjusted to include ∆G and product inhibition. Students will then measure population dynamics, yield and methane production of mixed cultures of degraders and methanogens. Different initial ratios of degraders and methanogens will be tested to determine whether growth and methane production is maximized at some intermediate ratio of the two species.
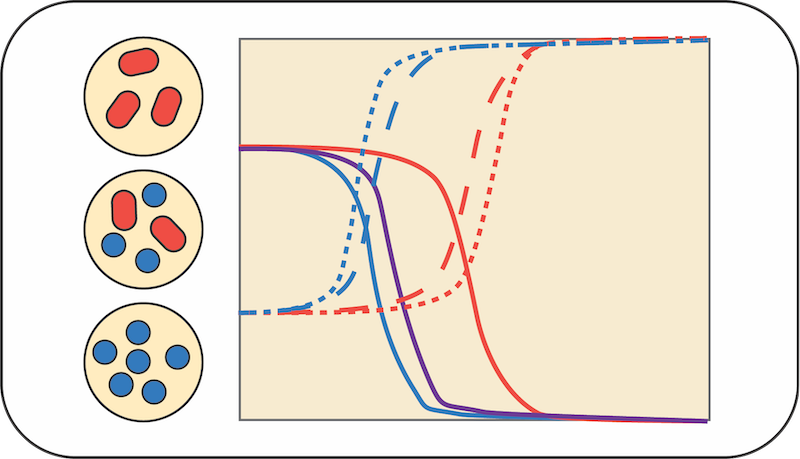
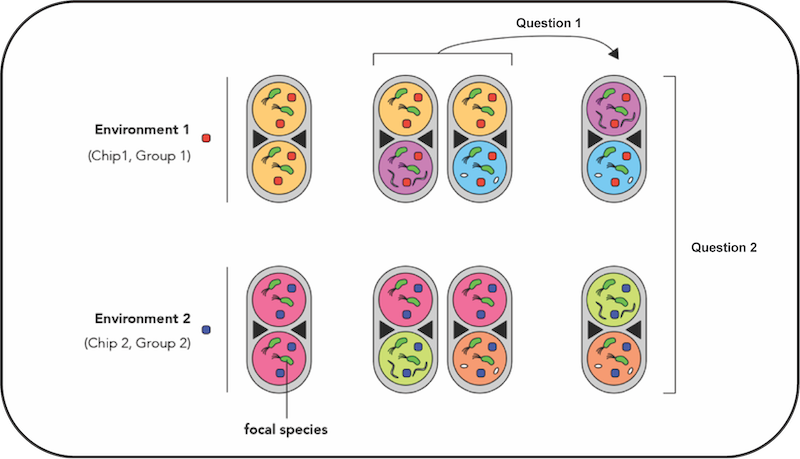
Droplet microfluidics for high-throughput microbial ecology
Jonathan Friedman, with Anthony Ortiz and Chandana Gopolakrishnappa
The development of new technologies has enhanced our ability to measure species interactions and elucidate rules of community assembly. In this module, students will learn about kChip technology, which enables the rapid construction and screening of parallel microbial communities generated from libraries of microbial monocultures. Students will design simple experiments to generate thousands of one-, two-, and three-species communities on different carbon sources. The data generated will be used to address two key questions: 1) Can we predict the yield of a focal species in co-culture with two species given its monoculture yield and yield with all individual species? 2) How well do predictive models for yield of focal species generalize across different environments? Students will generate and test different mathematical models for interspecies interactions and explore how these models can be improved such that they generalize more broadly to different environments.
Competition dynamics and disease outcome in the microbiome
Britt Koskella and Mike Boots
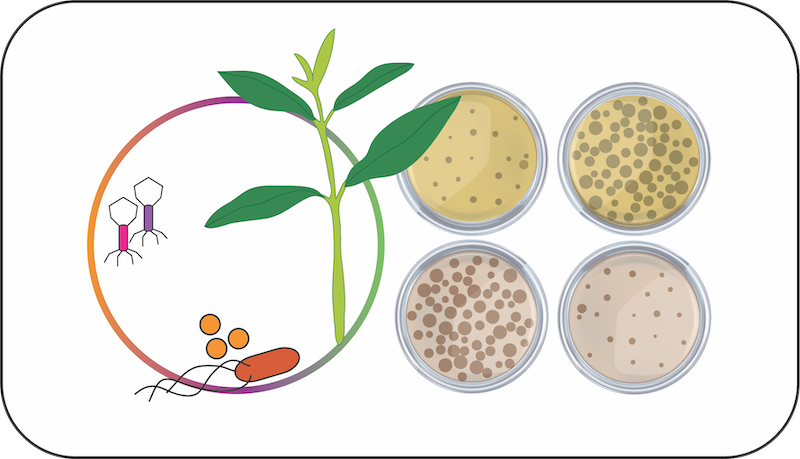
There is renewed interest in phage therapy to treat disease, yet the success of phage therapy is dependent on the context of the host microbiome in which they are applied. This module will investigate whether bacteriophage viruses influence the colonization success of bacterial pathogens by altering competition dynamics in the microbiome. Students will learn how to apply system-specific modeling approaches to predict and test these dynamics. Students will first measure quantitative phage resistance (i.e. the degree to which a given phage impacts the growth of a particular host) across plant-commensal and pathogenic strains of Pseudomonas. This data will be used to parameterize simple models of how apparent competition among strains might impact pathogen growth and disease. We will then test these predictions both in vitro and in planta by inoculating replicate microcosms and seedlings with pairs of commensal/pathogen species and a shared phage with differing plaquing efficiency on each. The work will act as proof of concept for the development of ‘trojan phages’ to reduce the colonization potential of bacterial pathogens.
Dynamics of microbial ecosystems in sediment
Alex Petroff
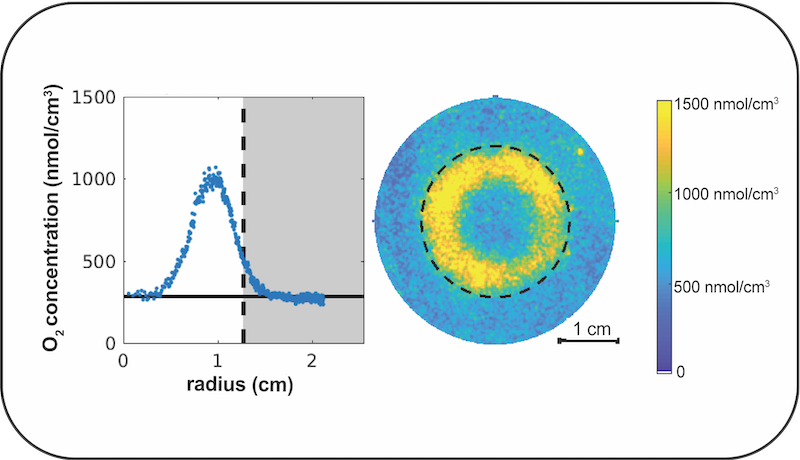
Microbial ecosystems living in sediment provide a microcosm in which to study nutrient cycling dynamics. Microbes in these ecosystems are dense and diverse: a handful of sediment contains billions of individual bacteria constituting millions of unique genotypes. The complementary metabolisms of sediment microbes produce oxygen, carbon, sulfur, nitrogen, and phosphorus gradients and allow these chemicals to be internally cycled. Since transport through sediment pore space is dominated by diffusion, the spatial scale of these gradients is small, typically millimeter scale. These dynamics can be faithfully modeled using systems of reaction-diffusion equations, and the small scale allows us to observe metabolic dynamics in tabletop experiments. This module will introduce students to the structure and dynamics of sediment ecosystems and techniques to measure, interpret and model the dynamics of oxygen gradients in sediment. Students will learn how to make and use planar oxygen optodes. These oxygen detectors are composed of a plastic sheet impregnated with a dye (Ru-dpp), the fluorescence of which is reversibly quenched by oxygen. The spatial distribution of oxygen can be measured by illuminating the optode with blue light. Next, students will build growth chambers that allow the continuous measurement of oxygen gradients in sediment while controlling the illumination, temperature, and atmospheric composition. Students will perform experiments to measure how oxygen gradients evolve as microbes grow, die, and move. Finally, students will learn how to invert the measured oxygen concentration for the local rates at which oxygen in produced and consumed.