
Application deadline: February 26, 2023
Course Subject:
Animal morphogenesis, the question of how body parts acquire their form, remains a fascinating open problem of developmental biology and physics. Molecular analysis uncovered how the basic architecture of the body plan is laid out and how genetic patterning instructs cell fate. Quantitative investigation of cellular flow in vivo indicates that feedback mechanisms between genetics, mechanical force generation, and geometry ensure a robust and reproducible time course of shape. These analyses rely on studying embryogenesis in model organisms such as the fruit fly, mouse, or zebrafish. While superb genetic perturbations are readily available, physical analysis, such as measurements or even perturbation of mechanics, remains a high hurdle.
Cell biology successfully incorporated experimental methods from physics and engineering into the study of immortalized cell lines. Culturing techniques enable precise regulation and measurements of biochemical factors, as well as physical, and geometric constraints. Recent progress with stem cell culture now enables addressing fundamental questions of developmental biology, about the interplay of mechanics, biochemical patterning, and geometry. This course will study synthetic morphogenesis by integrating quantitative data analysis and mathematical modeling, in a variety of hands-on projects. Experimental systems include the mechanobiology of human embryonic stem cell patterning, mechanical pattern formation in the avian dermis, synthetic morphogenesis with mouse as well as human embryonic stem cell lines, and the regenerative model system hydra.
Course Directors:
Instructors:
Idse Heemskerk (Univ. of Michigan), Kinneret Keren (Technion), Alan Rodrigues (Rockefeller ), Amy Shyer (Rockefeller), and Vikas Trivedi (EMBL Barcelona)
Guest Lecturers (preliminary list):
The summer course is closely linked to the concurrent KITP program Dynamics of Self-Organization in Animal and Plant Development. Course participants will attend the program's daily research seminars as part of the course curriculum. Students and lecturers will also have frequent opportunities for less formal interactions. Confirmed program participants include Dominique Bergmann (Stanford), James Briscoe (Crick Inst.), Francis Corson (ENS Paris), Cassandra Extavour (Harvard), Ken Irvine (Rutgers), Thomas Lecuit (College de France/IBDM), Ani Kicheva (IST Austria), Prisca Liberali (FMI), Adam Martin (MIT), Elliot Meyerowitz (Caltech), Rashmi Priya (Crick Inst.), Sharad Ramanathan (Harvard), Jochen Rink (MPI-NAT), Adrienne Roeder (Cornell), Timothy Saunders (Warwick), Eric Siggia (Rockefeller), Katharina Sonnen (Hubrecht Inst.), David Sprinzak (Tel Aviv), and Aryeh Warmflash (Rice).
Course Structure
Students will attend Dynamics of Self-Organization in Animal and Plant Development program talks each morning and spend afternoons and evenings working in their lab groups and attending modeling and analysis tutorials. Interaction with program speakers will also be facilitated through tutorial sessions and informal meetings over coffee and meals, as well as social evenings held jointly with the program. Students will be matched to experimental modules based on their ranked choices and have ample room to explore their own interests, based on initial instructor-guided projects. Students in different modules will interact informally – many research areas benefit from comparison or use similar technical approaches. Students teach each other, sharing their different expertise--project groups will intentionally include students from different academic backgrounds--and work closely with expert instructors, TAs. and guest lecturers from the associated KITP program.
There is no course fee, and students are provided with housing at UCSB's Manzanita Village residence halls and a 19-meal plan at campus dining commons. A typical day's schedule includes:
- 2 morning talks at KITP
- Lunch in a dining hall or campus cafe, with other course or program participants
- Afternoon lab work
- Dinner in a dining hall, followed by evening lab work and/or science discussions
On weekends, students can attend program BBQs and self-organize outings to the beach or around Santa Barbara.
Experimental Projects (partial list)
Amy Shyer and Alan Rodrigues: Morphological symmetry breaking in self-organizing mesenchymal tissue
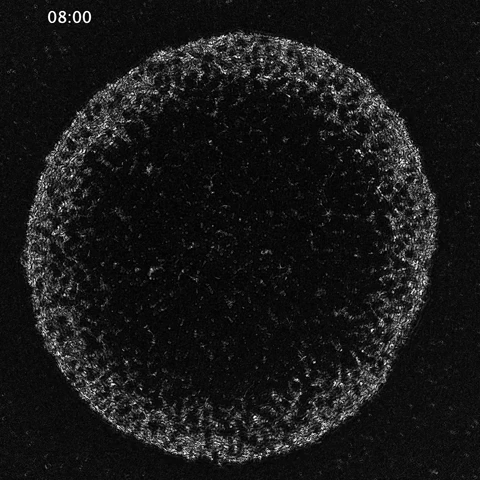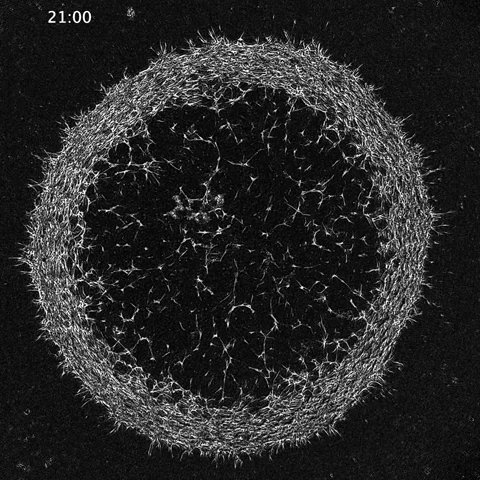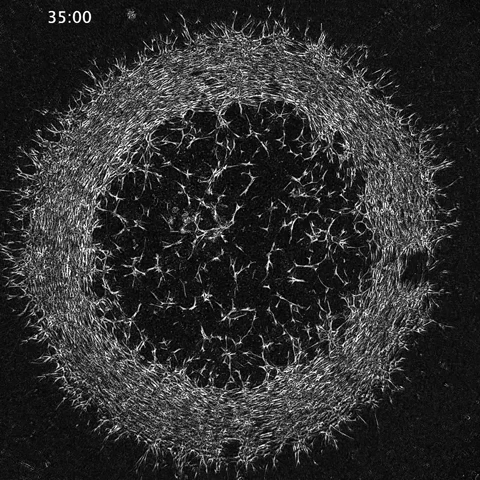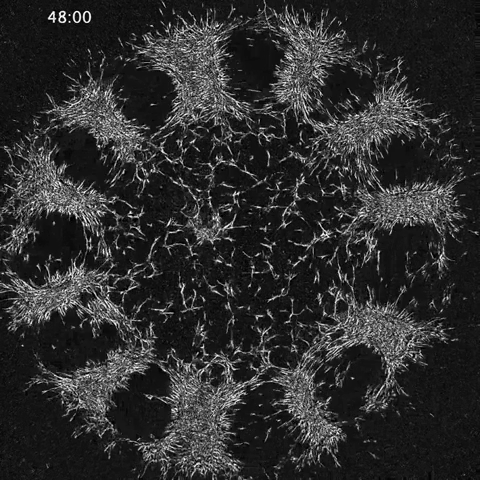
While much of the recent advances in our understanding of mechanics in development have focused on epithelial tissues, the Shyer-Rodrigues Laboratory of Morphogenesis studies morphological symmetry breaking driven by self-organizing mesenchymal tissues. Using a first-of-its-kind experimental platform, we combine live-cell imaging with collagen substrates specifically engineered to enable skin progenitor cells to self-organize into follicle patterns (https://www.quantamagazine.org/embryo-cells-set-patterns-for-growth-by-pushing-and-pulling-20220712/). By recapitulating a dynamic in vivo process, we will observe and manipulate multicellular activity in a tissue as it is generated.
___________________________________________________________________________________________________________
Idse Heemskerk: Predicting cell fate in pluripotent stem cells
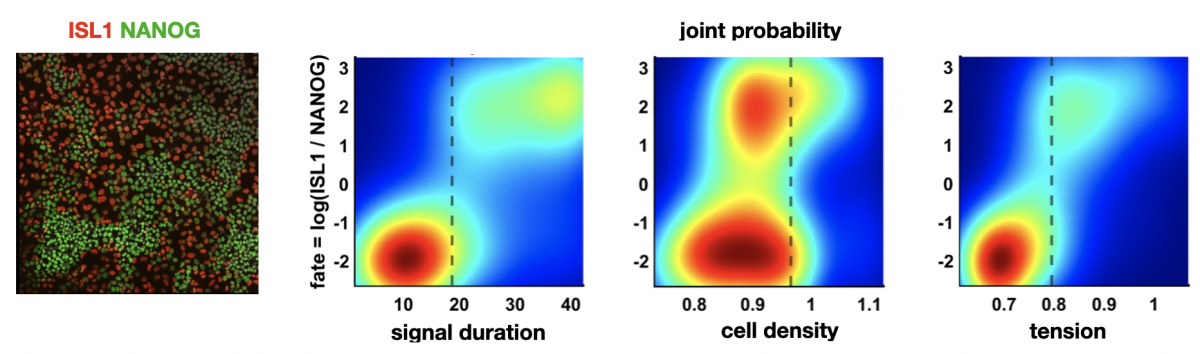 Figure: left: cells stained for fate markers after 42h of differentiation, right: (partially hypothetical) probability distributions showing how different factors relate to fate
Figure: left: cells stained for fate markers after 42h of differentiation, right: (partially hypothetical) probability distributions showing how different factors relate to fate
Pluripotent stem cells have remarkable capacity to self-organize into heterogeneous structures consisting of multiple cell types. Quantitative understanding of what makes one cell differentiate to one fate and a neighboring cell to a different fate is in most cases lacking. Variations in initial cell state, differences in morphogen signaling, and the mechanical environment all contribute, but their relative contributions and interdependence are often difficult to dissect.
In the context of the first cell fate decisions after pluripotency loss, associated with gastrulation, we will determine using single cell analysis of live and fixed imaging data how much redundant and non-redundant information different factors contain about cell fate. For example: does cell geometry at the onset of differentiation predict fate? What about the morphogen signaling dynamics? If so, does geometry predict morphogen signaling dynamics or are they independent inputs? And how do these relationships depend on substrate stiffness and intercellular forces?
Left figure: cells stained for fate markers after 42h of differentiation. Right figures: (partially hypothetical) probability distributions showing how different factors relate to fate. Figures by Seth Teague, unpublished.
___________________________________________________________________________________________________________
Kinneret Keren: Cytoskeletal dynamics in regenerating Hydra
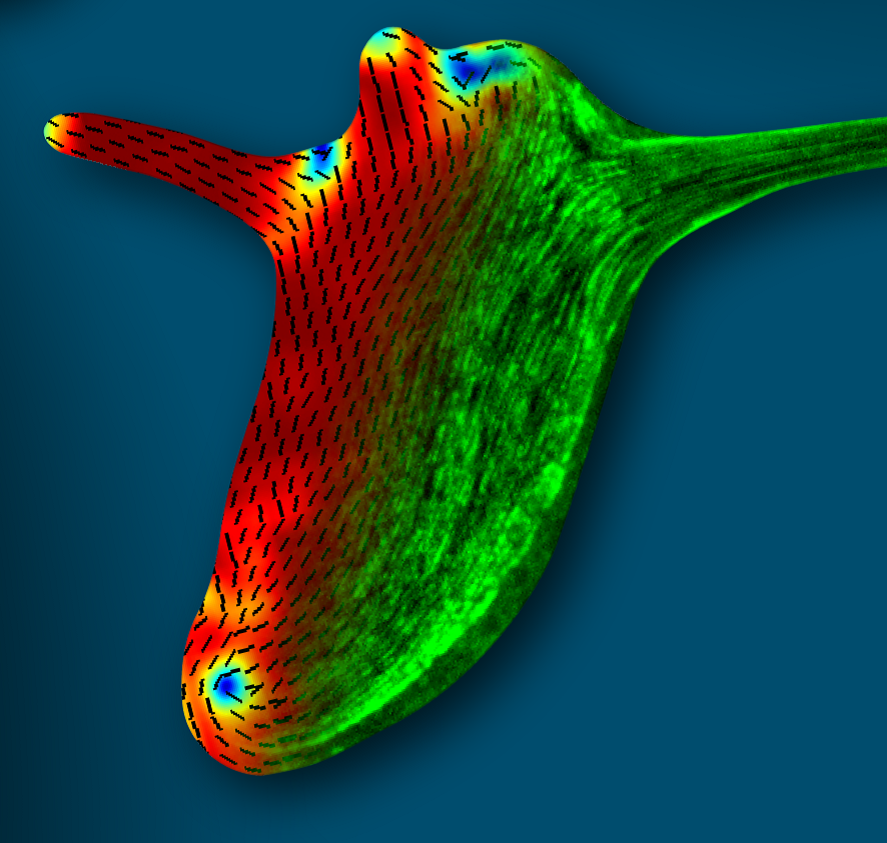 Figure: The supracellular actin fibers in a small regenerated Hydra (green) plotted together with the orientation field (black lines) and the nematic order parameter (jet colors).
Figure: The supracellular actin fibers in a small regenerated Hydra (green) plotted together with the orientation field (black lines) and the nematic order parameter (jet colors).
Hydra is a small fresh-water animal renowned for its extraordinary whole-body regeneration capabilities. We will study the emergence of large-scale order in the cytoskeleton of regenerating Hydra, focusing on the establishment of parallel arrays of supracellular contractile actomyosin fibers. We will excise tissue pieces from transgenic Hydra expressing lifeact-GFP in the ectoderm and/or endoderm, and follow the dynamic organization of the actin cytoskeleton during regeneration using live microscopy. We are particularly interested in understanding how the large-scale nematic order develops in orthogonal directions in the two epithelial tissue layers, and how this relates to the morphogenesis process.
___________________________________________________________________________________________________________
Vikas Trivedi: Biophysics and metabolism in self-organising structures from embryonic stem cells
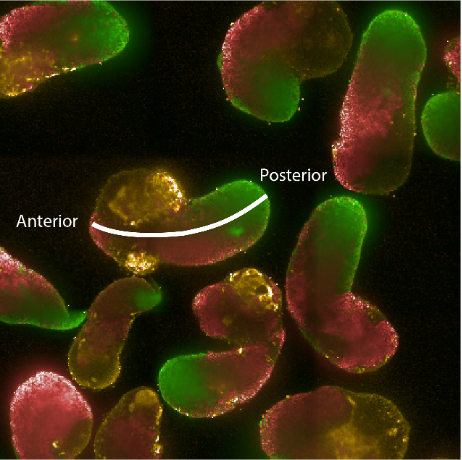 Figure: Hybridisation chain reaction (HCR) staining of 96h gastruloids. RNA expression patterns of Brachyury (green - primitive streak/mesodermal), Sox2 (red - pluripotent/ectodermal) and Sox17 (yellow - endodermal). The white line indicates the anterior-posterior axis illustrating the spatial organisation of cell fates in the gastruloid model.
Figure: Hybridisation chain reaction (HCR) staining of 96h gastruloids. RNA expression patterns of Brachyury (green - primitive streak/mesodermal), Sox2 (red - pluripotent/ectodermal) and Sox17 (yellow - endodermal). The white line indicates the anterior-posterior axis illustrating the spatial organisation of cell fates in the gastruloid model.
How can tissue shapes and patterns emerge reproducibly and robustly in multicellular systems like animals? Despite more than 100 years of embryology, it still remains unclear how gene networks, forces and mechanical properties integrate with the metabolic state of the cells to self-organize complex structures. This is due to our inability to disentangle the combined action of these factors (biophysical properties, gene networks and metabolic activity) within populations of genetically equivalent cells. For example, the activity of genetic programs is accompanied by emergence of distinct biophysical characteristics within different cell types. At the same time the metabolic state of a cell can influence cell fate decisions by modifying transcription factor signalling, the epigenetic landscape as well as cellular mechanics. Making quantitative measurements and systematic perturbations of these factors are nearly impossible in vivo. Therefore, we will take advantage of aggregates of mouse embryonic stem cells that recapitulate hallmarks of early embryonic development in vitro. This will allow us to explore the role of metabolism in symmetry breaking, germ layer specification and emergence of distinct biophysical properties that mould the tissue. We will use machine-learning based image analysis, quantitative measurements of relative material properties and genetically engineered cells with live imaging to dissect how coordinated changes in metabolic states and biophysical properties affect morphogenesis.
___________________________________________________________________________________________________________
Idse Heemskerk, Sebastian Streichan and Xavi Trepat: Linking fate and force in micro patterned human pluripotent stem cells
Micropatterned colonies of human pluripotent stem cells (hPSCs) recapitulate aspects of human embryonic development. Such systems provide an opportunity to study developmental events that are otherwise inaccessible to quantitative experimental approaches. This cell culture environment offers new interrogation and perturbation approaches that promise insight into the dynamics early human morphogenesis. These include spatial force measurement maps, tuning substrate stiffness, and shape as well as size of the micropatterned hPSCs colonies. We seek to exploit these advantages and leverage synergies between groups to study the interplay of biochemical signaling, tissue mechanics, and geometry. Specifically, we will have a project studying how the interplay between secreted factors and mechanical forces drives the formation of primordial germ cells in humans. Another example will investigate how mechanical stresses are coordinated to achieve neural plate folding into a tube.
