
Application deadline February 1, 2026
Course Subject:
Morphogenesis is the process by which cell growth, division, and differentiation are tightly controlled to form properly shaped, functioning organs. This remarkable control over form, fate, and function is coordinated by a tight interplay of the genetic program of development and physical interactions between cells. How do living organisms reliably build complex structures from single cells, despite the variability of their components and environments? What determines the emergence of organized patterns, the timing of developmental transitions, or the reproducibility of shape across individuals and species? While genetics has revealed many of the molecular players involved, the fundamental principles that govern the transformation from cell to tissue to organism remain elusive. Developmental systems exhibit remarkable coordination across scales—from gene expression to mechanical forces to tissue geometry—but how these levels interact to produce robust morphogenesis is still not well understood. This hands-on research course will integrate laboratory projects, computational data analysis, and mathematical modeling, aiming to achieve quantitative understanding of specific morphogenetic processes in a variety of model animal and plant developmental systems.
Course Directors:
Instructors:
Guest Lecturers (partial list):
The summer course is closely linked to the concurrent KITP program Geometry and Intercellular Interactions in Morphogenesis of Animals and Plants. Course participants will attend the program's daily research seminars as part of the course curriculum. Students and lecturers will also have frequent opportunities for less formal interactions. Confirmed program participants include Dominique Bergmann (Stanford), Fridtjof Brauns (MPI-PKS), James Briscoe (Crick Inst.), Danelle Devenport (Princeton), Stefano Di Talia (Duke Univ.), Madhav Mani (Northwestern), Elliot Meyerowitz (Caltech), Anne-Lise Routier-Kierkzkowska (Univ. de Montreal), and Xavier Trepat (IBEC).
Course Structure
The course is 5 weeks, beginning with a one-week bootcamp, followed by two 2-week lab sessions. Each session will consist of about four experimental projects led by instructors and TAs working closely with groups of 4-5 students. The daily routine involves two morning lectures and discussions, followed by lab work in the afternoons until late in the evenings. During the bootcamp, students will acquire skills needed to succeed in research projects, including microscopy, quantitative image analysis, and theoretical modeling. There will also be an introduction to model systems of development and laboratory techniques.
The morning lectures will take place in the context of the parallel running KITP program Geometry and Intercellular Interactions in Morphogenesis of Animals and Plants, providing an excellent setting for discussion and exchange with program participants. The scientific agenda of the program aims to closely study morphogenesis in animals and plants, bringing together these diverse communities with experts in physical modeling of morphogenesis.
The course will study how common physical challenges, experienced by animals and plants are solved. Plant and animal lineages diverged before the evolution of multicellularity, strikingly manifested at the molecular level, where each kingdom separately optimized genetic pathways.
Specifically, the course will seek to develop answers towards these questions:
- How are cell behaviors coordinated during development over long ranges?
- How are growing organs divided into distinct zones?
- Are there common organizing principles underpinning regulation of these processes across kingdoms?
- Is there mechanical regulation of growth?
A typical day's schedule includes:
- 2 morning talks at KITP
- Lunch in a dining hall or campus cafe, with other course or program participants
- Afternoon lab work
- Dinner in a dining hall, followed by evening lab work and/or science discussions
On weekends, students can attend program BBQs and self-organize outings to the beach or around Santa Barbara.
Accommodations, Fees and Financial Assistance
Students are housed in double-occupancy rooms in UCSB student apartments and provided with 19-meal plan at campus dining commons. The room and board fee is $2925. There are no other fees or tuition. Students who need financial assistance can request it in their course application form; financial need does not affect an applicant's chances of admission. A limited number of single rooms are available for a $1496 supplement.
Experimental Projects
Variation of early embryo compaction in echinoderm embryos
Instructors: Vanessa Barone* and Jana Sipkova
*Module Leader
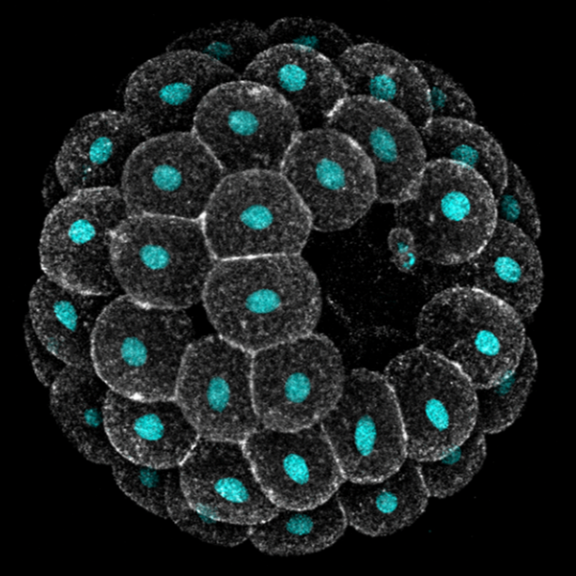
Embryos develop successfully only if they acquire a specific shape, i.e. species-specific geometry and 3D topology of embryonic cells. Yet, embryonic shapes present a certain degree of variation, which allows the generation of new life forms through evolution. What causes variation in embryonic shapes, what sets the limits to this variation, and why, remain open questions in developmental biology.
Echinoderms are excellent models to explore these questions: they produce billions of optically transparent embryos that develop freely in sea water, making them amenable to live imaging and embryology experiments. Moreover, the shape of early embryos is similar enough to allow meaningful comparison across species, and yet quite variable both within and between species.
In this module we will focus on variation in compaction of early embryos. Echinoderm zygotes are surrounded by a fertilization envelope and undergo holoblastic cleavage, giving rise to a mass of cells that organize into a monolayered epithelium encircling a cavity, i.e. a blastula. The way they do this, however, varies across species. Sea urchin cells immediately form a compact cluster that sits suspended in the middle of the fertilization envelope and subsequently opens a cavity in the middle. Sea star cells, instead, adhere to each other only loosely, line up on the inner surface of their fertilization envelope and progressively compact to form a tight epithelium. Previous work found that variation exists also between embryos of the same species: both the timing and extent of compaction is different among sibling sea star embryos, with implication for cell differentiation and survival.
Students will investigate the biophysical basis of this variation, with the goal of identifying the cellular and environmental factors that underlie differences in compaction dynamics between sea star embryos, and which ones of these factors sets the limits for what variation is observed in natural populations. Students will combine live imaging, embryological and biophysical methods to build a theoretical model for sea star compaction and its evolution. Depending on animal availability, these analyses will be extended to other echinoderm species, e.g. sea urchin and sea cucumber, with the goal of determining whether the same principles underlie inter-species variation.
Decoding Mechano-polarity Coupling During Plant Organ Growth
Instructors: Alexis Maizel* and Anamarija Primc
*Module leader
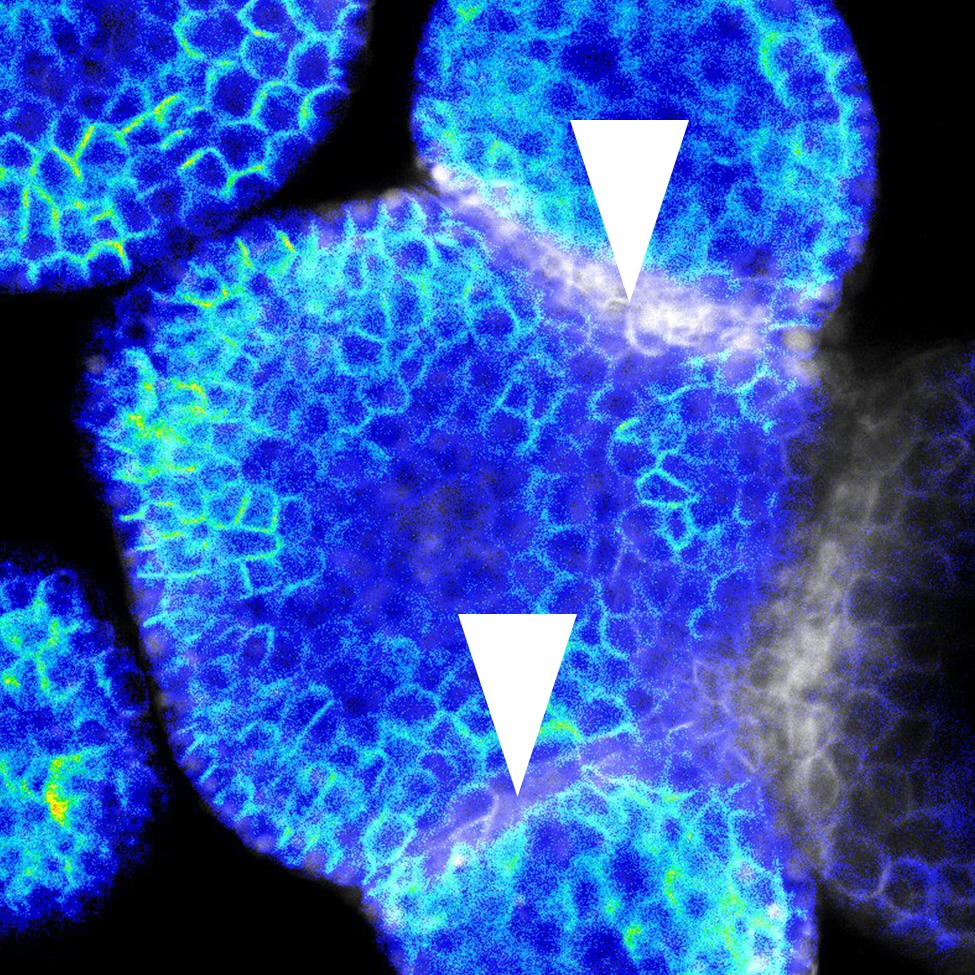
Plant organs self-organise robust shapes while cells experience heterogeneous growth and tissue-level mechanics. We lack a quantitative account of how mechanical state, signalling dynamics (e.g., auxin, cytokinin), and cell/tissue geometry each contribute to polarity establishment and division orientation --and how these inputs interact across organs and stages.Using single-cell analysis of live and fixed imaging across representative organs (root primordia, primary root, shoot apical meristem, leaf primordia), quantify the contribution of (i) local mechanics, (ii) signaling dynamics, and (iii) geometry/growth provide on polarity readouts and cell division orientation during organ growth.
Cell shapes and tissue geometry as a predictive framework for developmental robustness
Akankshi Munjal*, Kira Heikes, and Yusuke Mori
*Module leader
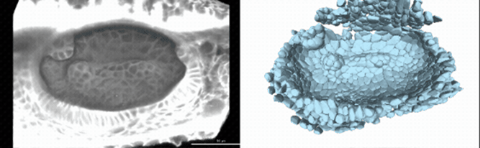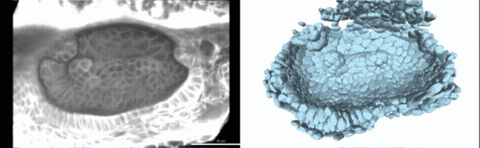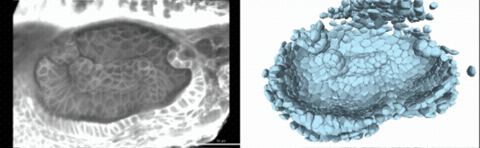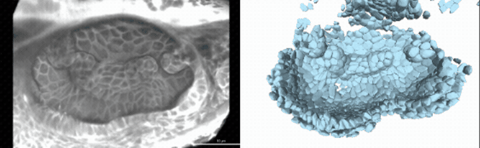
Using live imaging and quantitative shape analysis of zebrafish otic vesicles, we will explore how tissue geometry encodes developmental information. This simple, ovoid epithelial tissue undergoes dramatic remodeling to form the semicircular canals– three intricately shaped tubes that allow balance and spatial navigation. By applying genetic, environmental, and pharmacological perturbations, we will investigate how cell shapes and tissue geometry can serve as a predictive framework to understand when development proceeds robustly and when it fails."
Timelapse legend- "Timelapse of zebrafish canal morphogenesis: A 3D-rendered zebrafish otic vesicle at 2 days post fertilization visualized using a fluorescent membrane-labeled line (left) and its cell-segmented reconstruction generated with Cellpose–SAM (right)."
Amazingly elastic cuticle in honeypot ants: shape change and material properties
Naomi Nakayama* and Cedric Flint
*Module leader

Honeypot ants (genus Myrmecocystus) have specialized workers that consume large amounts of food to the point that their abdomens swell enormously — growing to about four to five times its normal dimension — and become transparent. Insect cuticle is mostly composed of chitin, pigments, and cuticular proteins that confer the cuticle its mechanical properties. It is reasonable to hypothesize that the presence of special cuticular proteins in the honeypot ant’s abdomen accounts for its exceptional elasticity. In this project, we will focus on the materials rather than the geometry itself. We will examine at various levels how elastic deformation can temporarily affect tissue geometry in adult organisms by integrating 3D microscopy, transcriptomics/proteomics data processing, and traditional material characterization tools. In addition, we will compare several pupal stages to determine when this superelasticity emerges during post-embryonic development. In parallel, we will evaluate the putative role of the elastomeric protein resilin using immunostaining as part of a candidate strategy. More generally, the drastic increase in volume raises questions about the potential structure-function of the ultimate ellipsoidal shape of the abdomen. Depending on the students’ interests and backgrounds, modeling could be used to address this topic.
Beyond mechanics: How tissue material phase transitions shape the embryo
Instructors: Nicoletta Petridou* and Laura Rustarazo Calvo
*Module leader
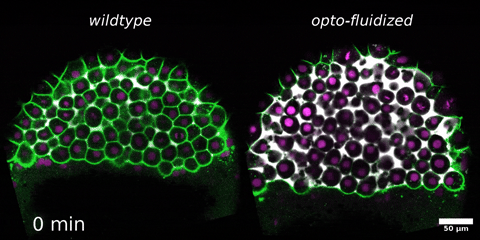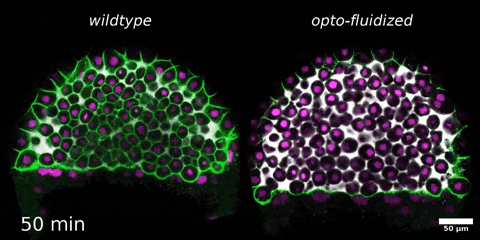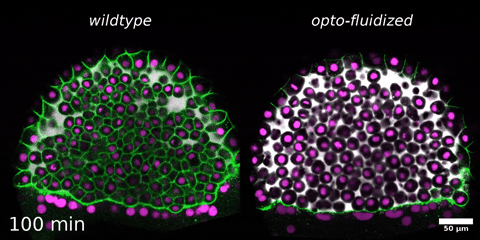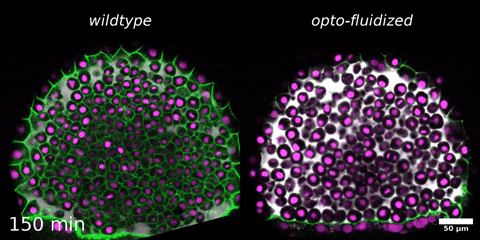
Morphogenesis relies on spatial and temporal patterns in tissue deformability, as well as on their abrupt changes. Such material phase transitions, e.g. between fluid- and solid-like states, occur when a microscale control parameter reaches a critical point. Cellular properties such as adhesion, density, and shape can each act as a control parameter of such transitions. In this module, we will experimentally engineer material phase transitions in the zebrafish pluripotent embryo by tuning the above control parameters and address how different types of phase transitions affect: i) tissue deformability, ii) tissue organization, iii) mechano-material sensation, and iv) signalling response. We will combine optogenetic perturbations, transplantations, live imaging of signalling reporters and cell dynamics, quantitative imaging analysis and phase transition theoretical frameworks. By the end of the module, participants will get a flavour of the rich role of collective tissue physical properties on decision-making during multicellular development.
Decoding the force: dynamic mechanical assays in a minimal model of human gastrulation
Instructors: Diana Pinheiro* and Chloé Roffay
*Module leader
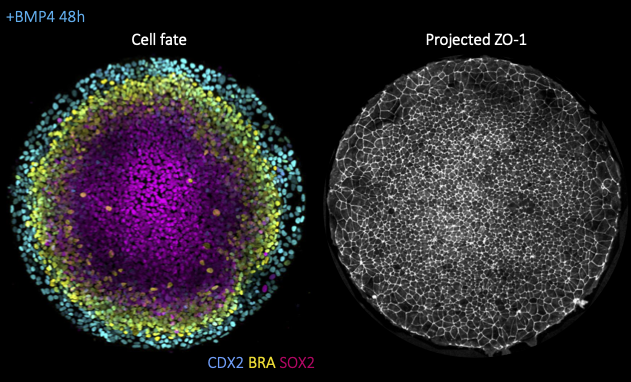
Alongside morphogen signaling, mechanical forces are emerging as important regulators of cell fate. This can result from indirect modulation of cell and tissue geometry, as previously shown for gut morphogenesis, from direct mechanosensitive activation of key developmental pathways (e.g. Wnt, YAP/TAZ), ion channels (e.g. Piezo) or from mechanically induced changes to chromatin architecture. While most studies focused thus far on the response to constant mechanical cues (e.g. substrate stiffness), it remains poorly understood how the magnitude versus duration of mechanical stress modulates the activity of mechanosensitive pathways and, ultimately cell fate. To tackle this question, we will take advantage of a minimal model for human gastrulation termed gastruloid discs. These are micropatterned human embryonic stem colonies which self-organize into the 3 germ layers (mesoderm, endoderm, ectoderm) and the extraembryonic amnion upon stimulation with BM4 ligands (left fig.). Using this system, we will then combine dynamic mechanical manipulations, namely stretch or compression assays, with high-resolution live imaging (fig. right), automated morphometric analyses and live-cell signaling/cell fate reporters
Leaf morphogenesis: how cells coordinate growth to maintain flatness
Adrienne Roeder* and Mauar Zimmermann
*Module leader
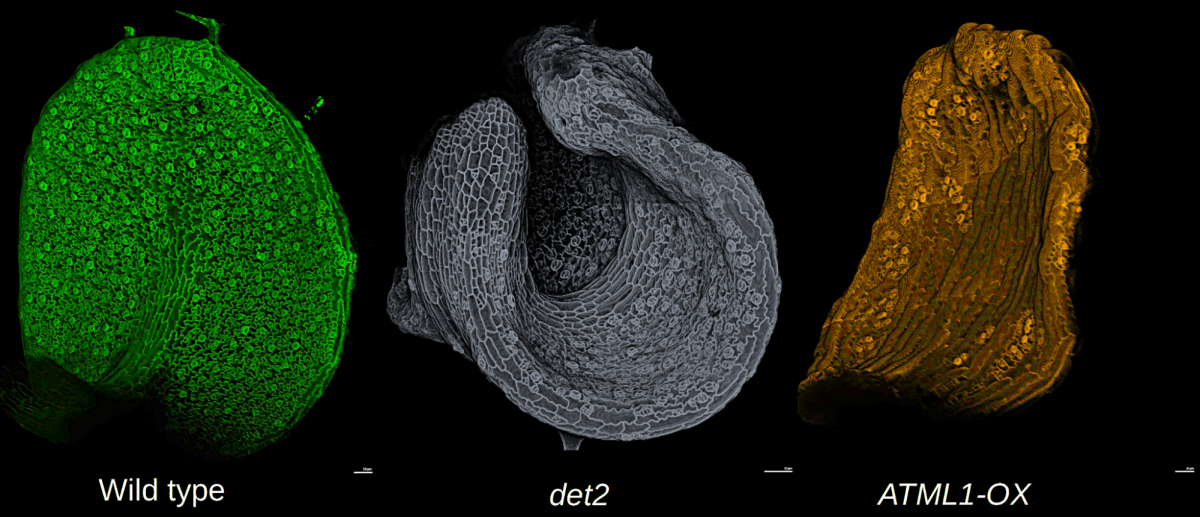
Have you ever tried to stretch a plastic bag? If so, you know it is nearly impossible to keep the plastic flat while it is rapidly expanding, yet leaves remain nearly flat during huge amounts of growth. How do the cells coordinate their growth to maintain leaf flatness? We will image living, growing Arabidopsis leaves of wild type and mutants that disrupt flatness. We will use computational analysis to address the multiscale problem of how cell growth is coordinated across the organ to control shape.
Quantifying Developmental Dynamics in Arabidopsis thaliana
Instructors: Paul Tarr* and Carla Evans
*Module leader
Plant shape and form are determined by the coordination of cell division rates, gene expression patterns, and cell wall properties. Arabidopsis thaliana has served as the model plant to study and elucidate the biological processes that govern plant development. Live-imaging of cell dynamics such as gene-expression patterns, hormonal regulation, and response to cell-autonomous and non-cell autonomous signaling dynamics have been essential tools for understanding plant development. In this course we will use live-imaging and methods to perturb developmental processes in a spatio-temporal manner to quantify changes in aspects of patterning, growth, and morphogenesis at the Arabidopsis shoot apical meristem, the above ground stem cell niche.
What is the KITP QBio Summer Research Course?
Understanding fundamental problems in biology requires both advanced experiments and the quantitative/theoretical approach traditionally taken by physics. The Santa Barbara Advanced School of Quantitative Biology “QBio” summer course trains early-career scientists to integrate these different approaches by having them do it all–they take part in novel, team-taught lab experiments led by renowned faculty; they integrate quantitative modeling and theory into their experimental design and data analysis; and they become part of an interdisciplinary community of biologists and physicists. The course aims to teach students to communicate and collaborate with people with different and complementary backgrounds: eliminating the scientific language barrier is a major goal of the course. Students leave the course with an expanded network of peers, senior program participants and course instructors. Ideally, students emerge from the course with much broader scientific horizons and confidence in their own ability to chart the path of their own research.
Who should apply?
The course is geared toward advanced graduate students and postdocs, whose rigorous disciplinary training and significant research experience has prepared them to absorb material through high-level seminars and a research environment. The high instructor-to-student ratio and team-based project format create a supportive environment, but the course is intensive and fast-paced. With only 5 weeks to work, students must be self-driven to get results. Since participants do much of their learning from teammates and through informal interactions with program participants, openness to others’ ideas (particularly those from different scientific and cultural backgrounds) is crucial, and the confidence to initiate conversations with senior scientists help successful students make the most of their time at KITP. Successful applicants typically show curiosity, a clear idea of how the course will impact their research program, a desire to do interdisciplinary work, and strong quantitative skills.
Last day #kitpqbio. After of 5 weeks of 24/7 work, students present their incredible achievements. Pure science! Then bbq at the legendary Munger residence. What a summer! Thanks @KITP_UCSB! pic.twitter.com/e5yq25re0W— Xavier Trepat (@XavierTrepat) August 25, 2023
What do students say about the course?
"Participating in research projects outside my PhD work and interacting with other researchers revealed that - (a) I am broadly curious about biology, (b) I can ask questions and formulate testable hypothesis and (c) I enjoy science. When one of the instructors admired my efforts in his project and offered me a postdoc position, I realized that my abilities to do science were greater than my perception and decided to develop them further by continuing in research." - Sapna Chhabra (2019)

“I think one of the greatest values of this course has been the ability to interact with people from different fields and be able to learn the jargon that they use in one field and understand it at a deep enough level that we can then have conversations and build on that. Being able to interact with the physicists, theorists, and other experimentalists has helped broaden my knowledge base and also made me more comfortable, as a student, being able to talk across different fields and learn from people doing very different types of research, but all interested in and centered around different kinds of questions.” - Jess Kanwal (2018)
“For me, the project was very helpful for understanding the gap between the idealized data we theorize about and the data we actually have, and the project and tutorials left me much more versed in common computational techniques for bacterial genomics. The morning talks and the connections built over many hours with the other students in lab and over meals was also inspiring and makes me more likely to continue pursuing an academic career.” - Michael McLaren (2017)

“The KITP/QBio summer course is quite unique in the world and has been a true accelerator for my career. It first gave me a broad, comprehensive and timely overview on the field of morphogenesis, from both physics and biology points of view. This has been critical to nurture my own research project and to successfully apply to group leader positions. As a theoretician, the school allowed me to perform my first biological experiments ever. I realized that 1) I was actually not too bad in doing experiments, 2) I’d probably very much like doing my own ones one day. . . . The practicals in small groups, mixing biologists and physicists was a fantastic opportunity to learn from each others and to gain better teaching skills. This was a perfect mix of theory and experiments, and a perfect setting to make emerge original research ideas.” - Herve Turlier (2016)
Spent amazing five weeks @KITP_UCSB during #morpho23 and #kitpqbio. I enjoyed the experimental work with "TEAM HANGRY", instructed by @viktri08 and @KStapornwongkul. Can't wait to be back @BauschLab to try new ideas, I got during discussions within this stimulating environment. pic.twitter.com/EbrF2KYGvW
— Marion Raich (@marion_raich) August 28, 2023
